15 Random forest
15.1 Practical experiments
15.1.1 Random forest for prediction of iris
The caret package (short for _C_lassification _A_nd _RE_gression _T_raining) is a set of functions that attempt to streamline the process for creating predictive models. The package contains tools for:
- data splitting
- pre-processing
- feature selection
- model tuning using resampling
- variable importance estimation
15.1.1.1 Required R packages
Required packages:
caret
AppliedPredictiveModeling
ellipse Attribute Information:
sepal length in cm
sepal width in cm
petal length in cm
petal width in cm
class:
– Iris Setosa – Iris Versicolour – Iris Virginica
str(iris)## 'data.frame': 150 obs. of 5 variables:
## $ Sepal.Length: num 5.1 4.9 4.7 4.6 5 5.4 4.6 5 4.4 4.9 ...
## $ Sepal.Width : num 3.5 3 3.2 3.1 3.6 3.9 3.4 3.4 2.9 3.1 ...
## $ Petal.Length: num 1.4 1.4 1.3 1.5 1.4 1.7 1.4 1.5 1.4 1.5 ...
## $ Petal.Width : num 0.2 0.2 0.2 0.2 0.2 0.4 0.3 0.2 0.2 0.1 ...
## $ Species : Factor w/ 3 levels "setosa","versicolor",..: 1 1 1 1 1 1 1 1 1 1 ...head(iris)## Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## 1 5.1 3.5 1.4 0.2 setosa
## 2 4.9 3.0 1.4 0.2 setosa
## 3 4.7 3.2 1.3 0.2 setosa
## 4 4.6 3.1 1.5 0.2 setosa
## 5 5.0 3.6 1.4 0.2 setosa
## 6 5.4 3.9 1.7 0.4 setosadim(iris)## [1] 150 515.1.1.2 Visulization
15.1.1.2.1 Scatterplot Matrix
#install.packages("AppliedPredictiveModeling")
library(AppliedPredictiveModeling)
transparentTheme(trans = .4)
#install.packages("caret")
library(caret)##
## Attaching package: 'caret'## The following object is masked from 'package:survival':
##
## clusterfeaturePlot(x = iris[, 1:4],
y = iris$Species,
plot = "pairs",
## Add a key at the top
auto.key = list(columns = 3))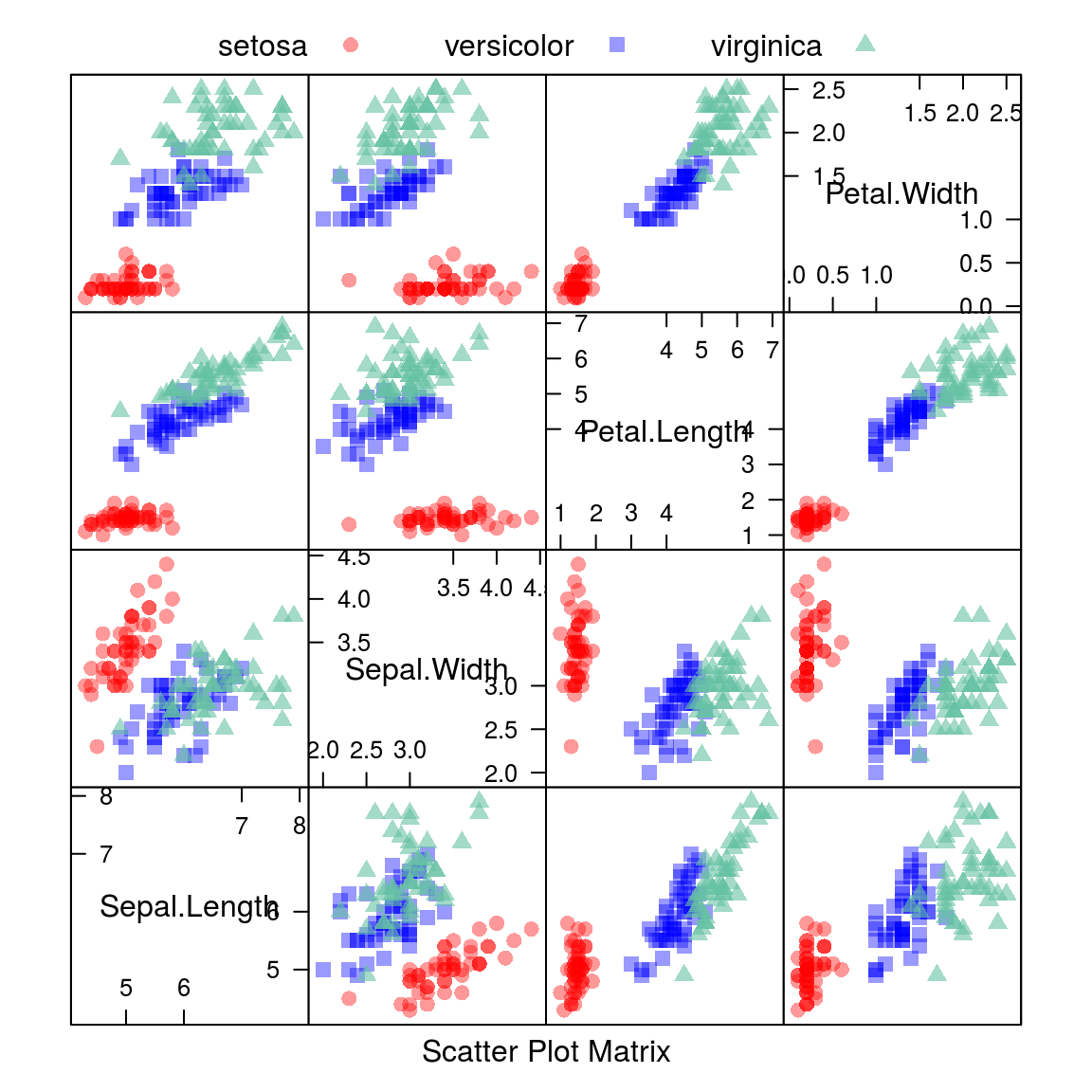
15.1.1.2.2 Scatterplot Matrix with Ellipses
#install.packages("ellipse")
library(ellipse)##
## Attaching package: 'ellipse'## The following object is masked from 'package:graphics':
##
## pairsfeaturePlot(x = iris[, 1:4],
y = iris$Species,
plot = "ellipse",
## Add a key at the top
auto.key = list(columns = 3))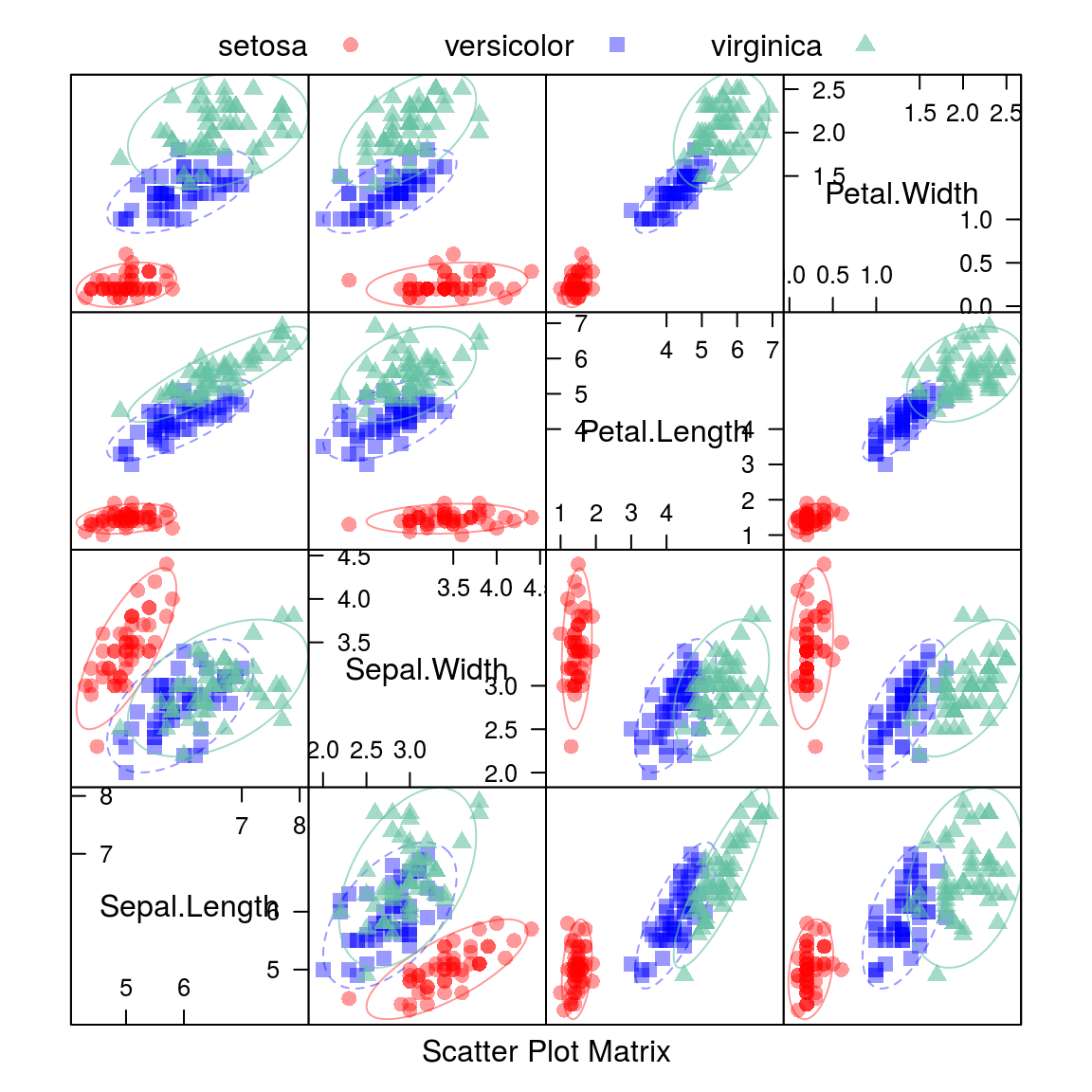
15.1.1.2.3 Overlayed Density Plots
featurePlot(x = iris[, 1:4],
y = iris$Species,
plot = "density",
## Pass in options to xyplot() to
## make it prettier
scales = list(x = list(relation="free"),
y = list(relation="free")),
adjust = 1.5,
pch = "|",
layout = c(4, 1),
auto.key = list(columns = 3))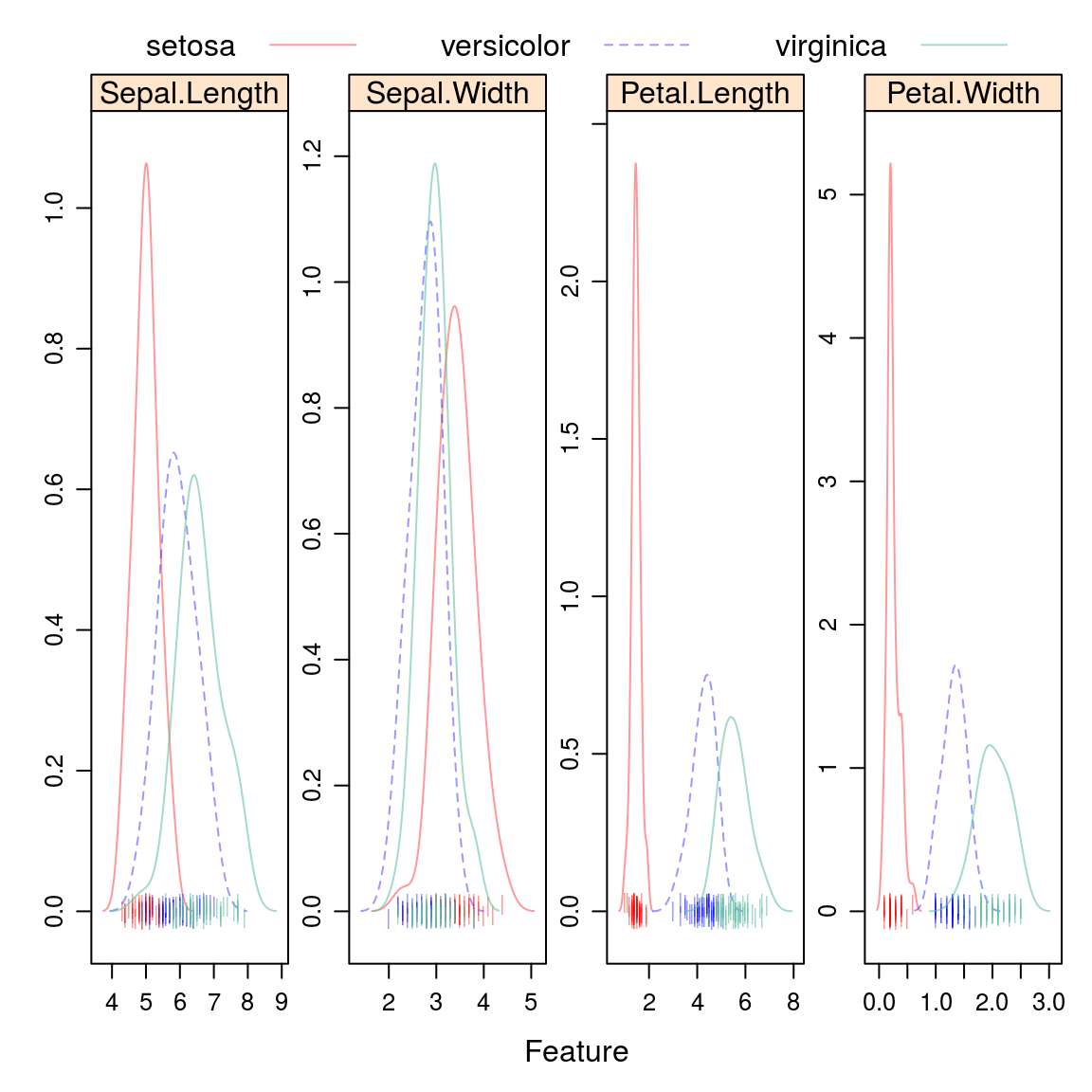
15.1.1.2.4 Box Plots
featurePlot(x = iris[, 1:4],
y = iris$Species,
plot = "box",
## Pass in options to bwplot()
scales = list(y = list(relation="free"),
x = list(rot = 90)),
layout = c(4,1 ),
auto.key = list(columns = 2))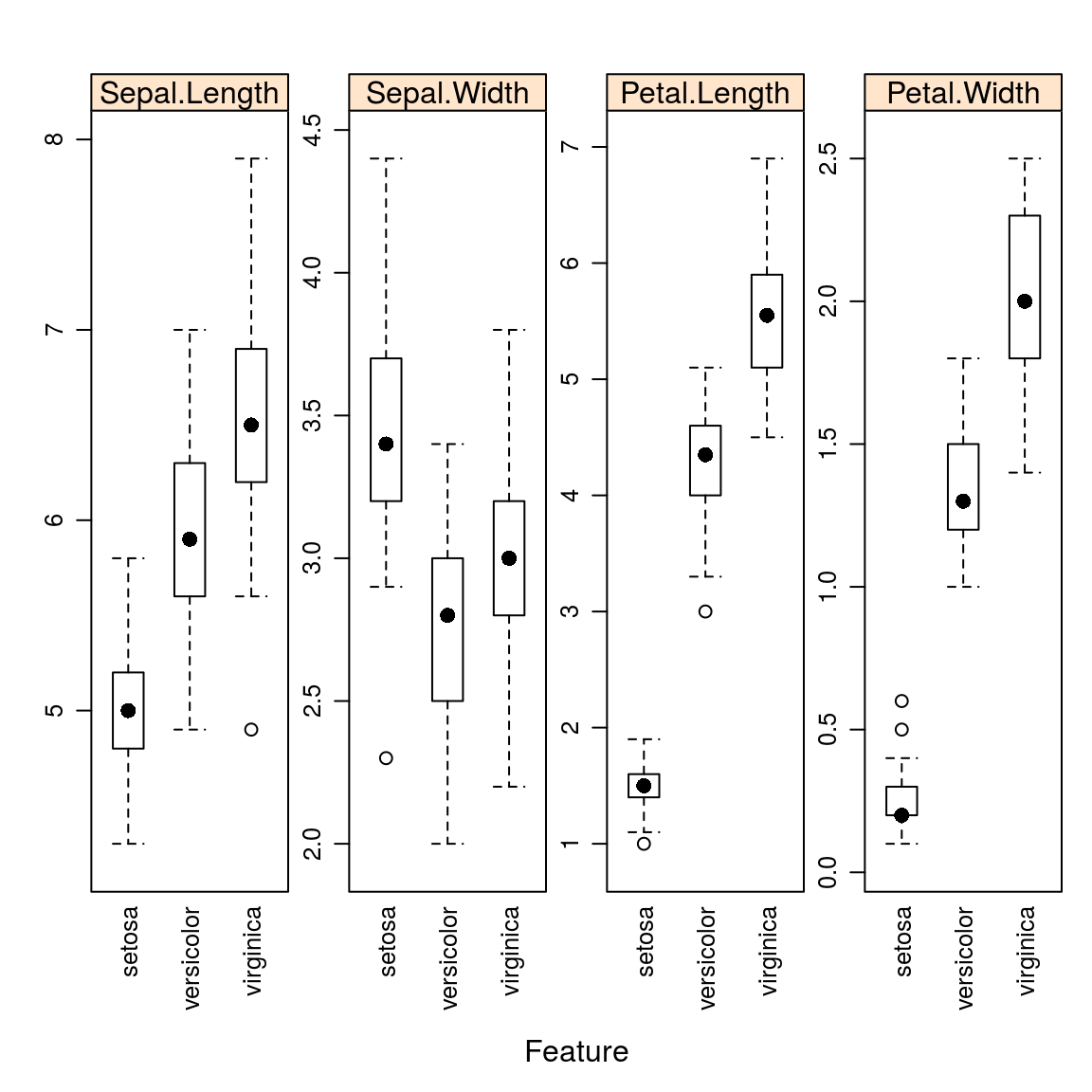
15.1.1.2.5 Machine leraning - Random forest
15.1.1.2.6 Model Train
set.seed(186)
# Data Splitting
train_index <- createDataPartition(iris$Species, p = 0.75, , times=1, list = FALSE)
train_set = iris[train_index, ]
test_set = iris[-train_index, ]
fit_rf_cv <- train(Species ~ ., data=train_set, method='rf', metric = "Accuracy",
trControl=trainControl(method="cv",number=5))
fit_rf_cv## Random Forest
##
## 114 samples
## 4 predictor
## 3 classes: 'setosa', 'versicolor', 'virginica'
##
## No pre-processing
## Resampling: Cross-Validated (5 fold)
## Summary of sample sizes: 91, 91, 93, 91, 90
## Resampling results across tuning parameters:
##
## mtry Accuracy Kappa
## 2 0.9398551 0.9100274
## 3 0.9398551 0.9100274
## 4 0.9398551 0.9100274
##
## Accuracy was used to select the optimal model using the largest value.
## The final value used for the model was mtry = 2.## Variance importance
rfVarImpcv = varImp(fit_rf_cv)
rfVarImpcv## rf variable importance
##
## Overall
## Petal.Width 100.00
## Petal.Length 95.11
## Sepal.Length 16.93
## Sepal.Width 0.0015.1.1.2.7 Testing test data
## predict
test_set$predict_rf <- predict(fit_rf_cv, test_set, "raw")
confusionMatrix(test_set$predict_rf, test_set$Species)## Confusion Matrix and Statistics
##
## Reference
## Prediction setosa versicolor virginica
## setosa 12 0 0
## versicolor 0 11 1
## virginica 0 1 11
##
## Overall Statistics
##
## Accuracy : 0.9444
## 95% CI : (0.8134, 0.9932)
## No Information Rate : 0.3333
## P-Value [Acc > NIR] : 1.728e-14
##
## Kappa : 0.9167
##
## Mcnemar's Test P-Value : NA
##
## Statistics by Class:
##
## Class: setosa Class: versicolor Class: virginica
## Sensitivity 1.0000 0.9167 0.9167
## Specificity 1.0000 0.9583 0.9583
## Pos Pred Value 1.0000 0.9167 0.9167
## Neg Pred Value 1.0000 0.9583 0.9583
## Prevalence 0.3333 0.3333 0.3333
## Detection Rate 0.3333 0.3056 0.3056
## Detection Prevalence 0.3333 0.3333 0.3333
## Balanced Accuracy 1.0000 0.9375 0.9375