Chapter 15 Heatmap Tutorial
15.1 Install pheatmap package
install.packages("pheatmap")15.2 Draw a heatmap for gene expression of RNA-seq data
library(pheatmap)
gene_exp <- read.table("data/maize_embryo_specific_gene_Sheet1.tsv", header=T, row.names=1)
pheatmap(gene_exp)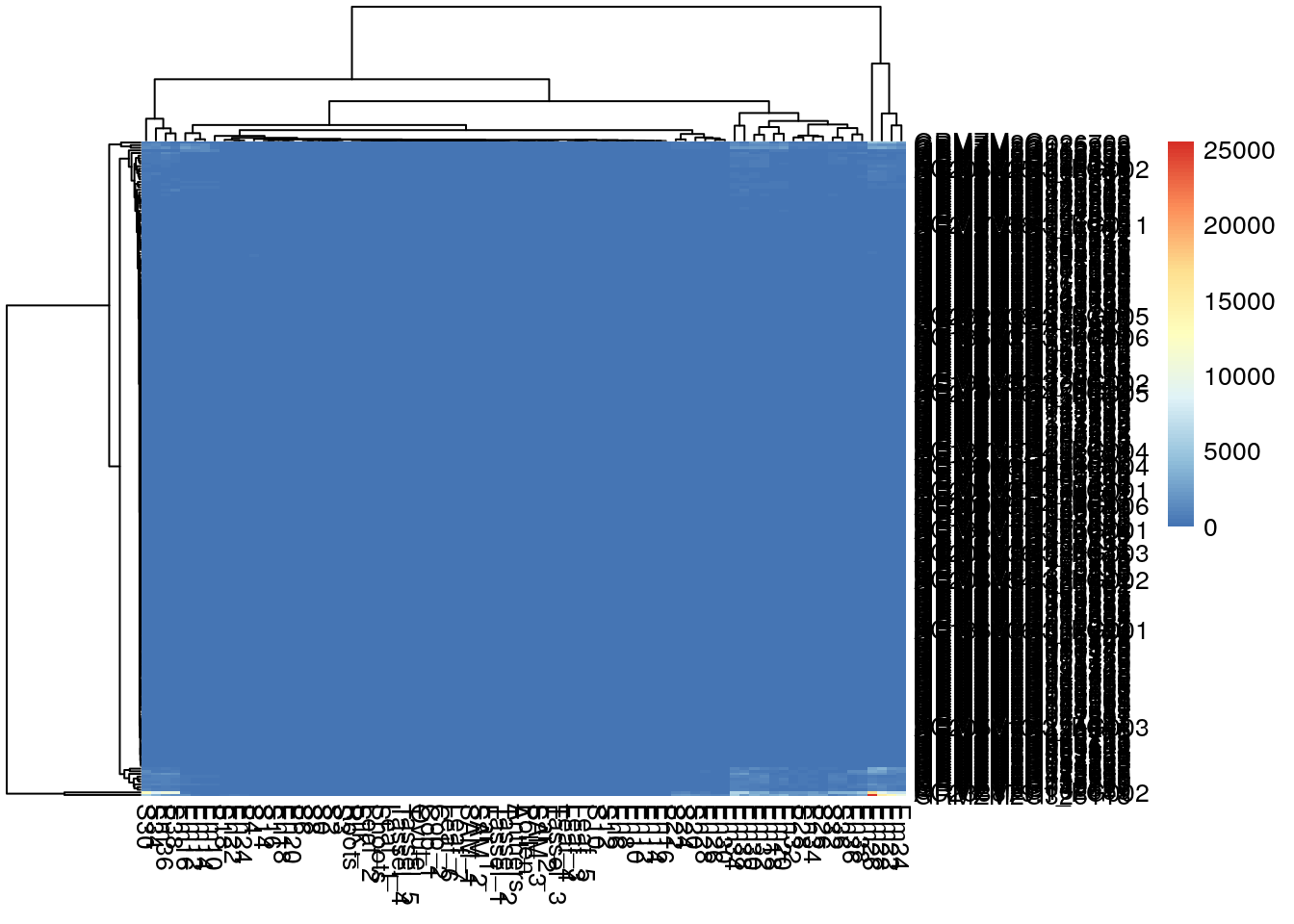
library(ggplot2)
# Set the theme for all the following plots.
theme_set(theme_bw(base_size = 16))
dat <- data.frame(values = as.numeric(unlist(gene_exp)))
summary(dat)## values
## Min. : 0.00
## 1st Qu.: 0.00
## Median : 0.40
## Mean : 38.91
## 3rd Qu.: 6.22
## Max. :25534.25ggplot(dat, aes(values)) + geom_density(bw = "SJ") + xlim(c(0,100))## Warning: Removed 958 rows containing non-finite values (stat_density).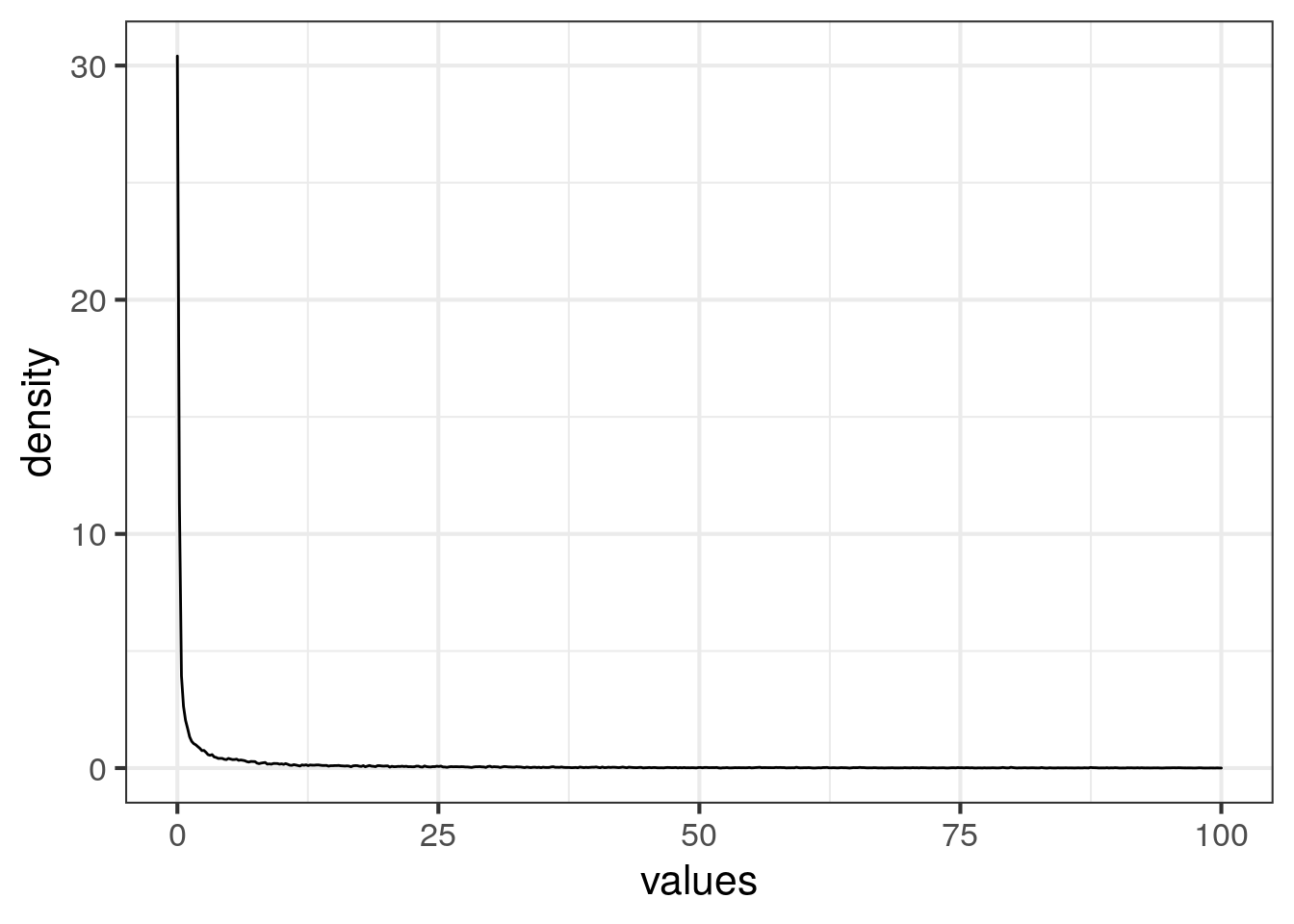
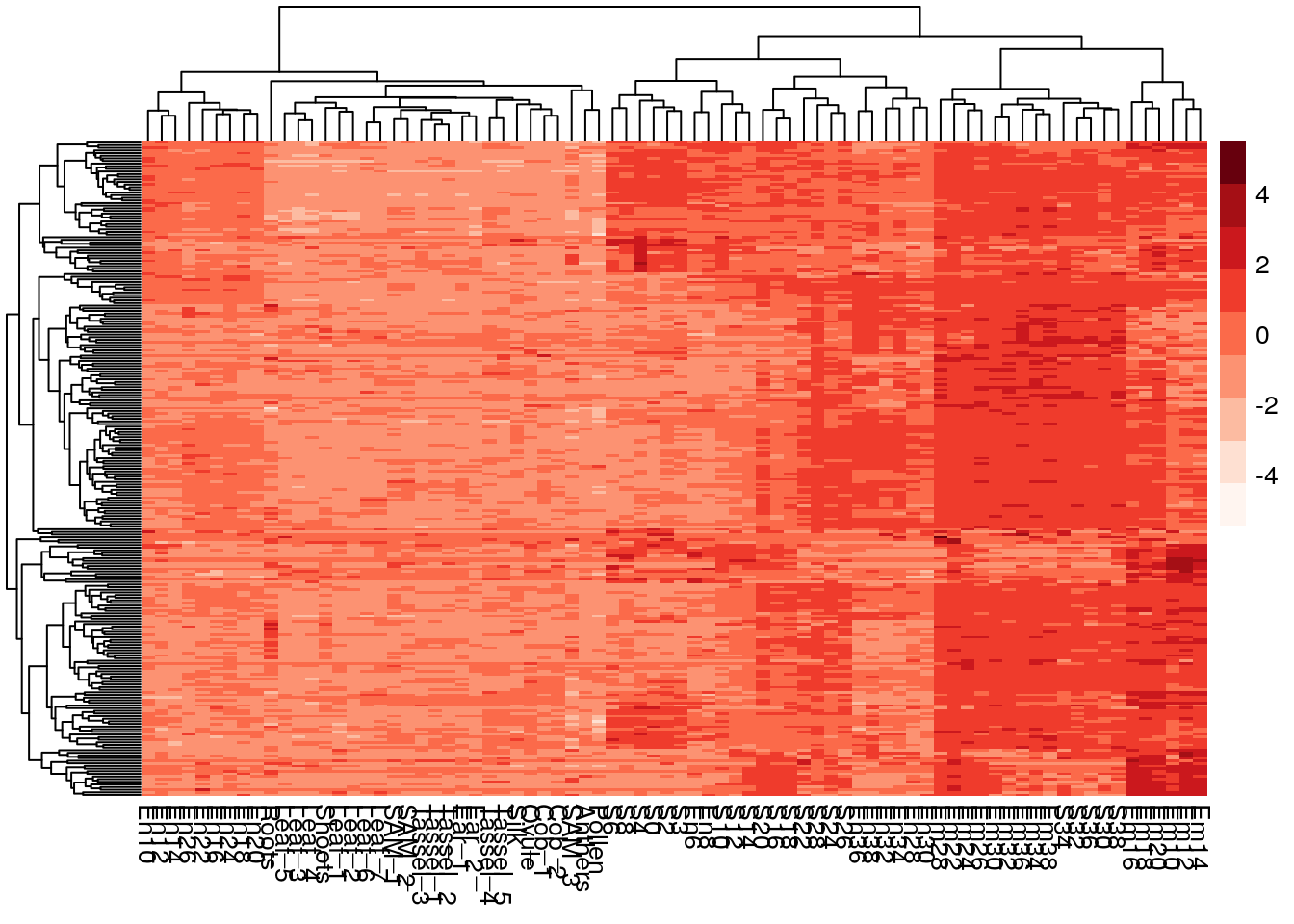
The data is skewed.
pheatmap(log2(gene_exp + 0.000001), scale="row")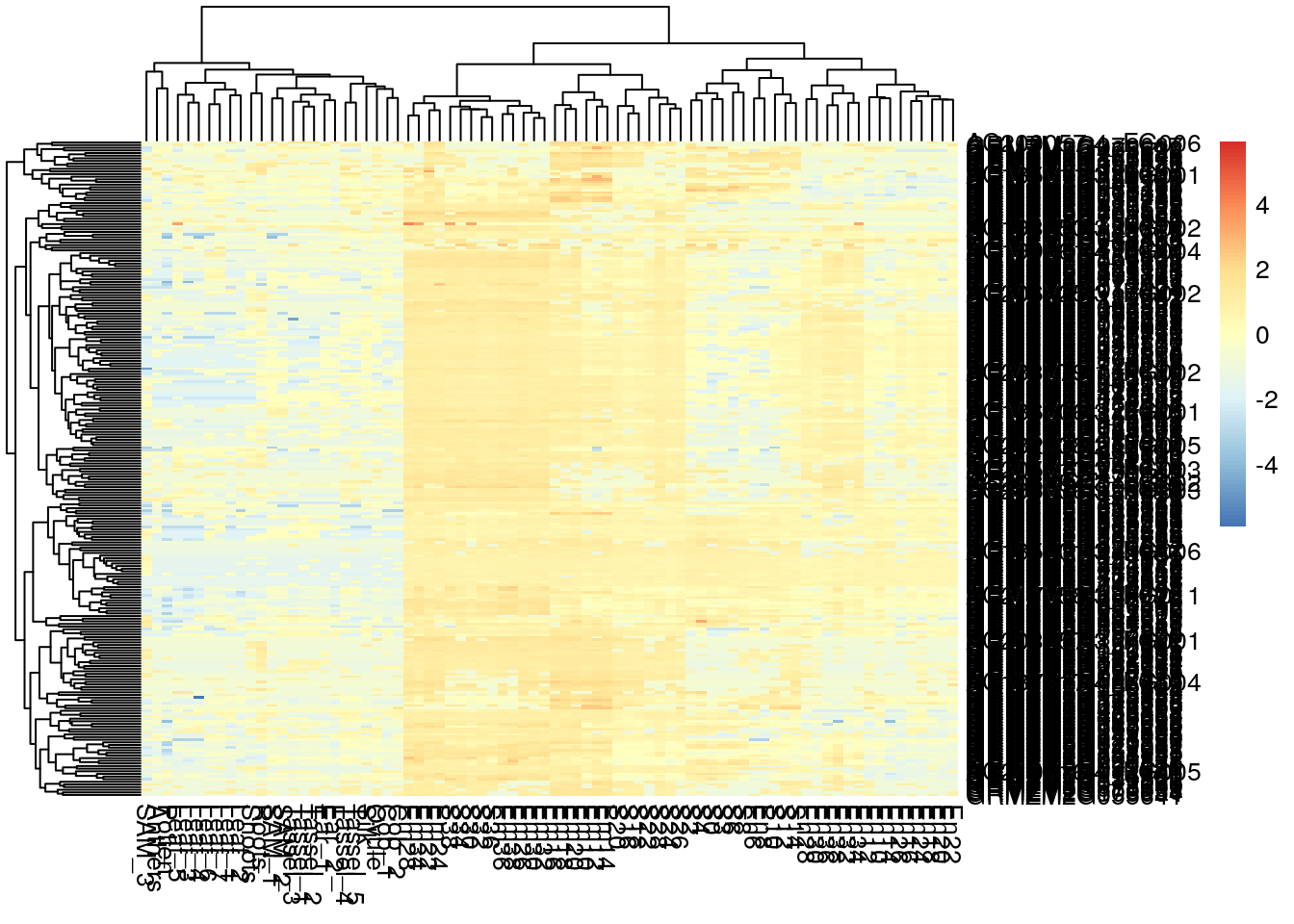
pheatmap(log2(gene_exp + 0.01), scale="row", show_rownames = T, show_colnames = F)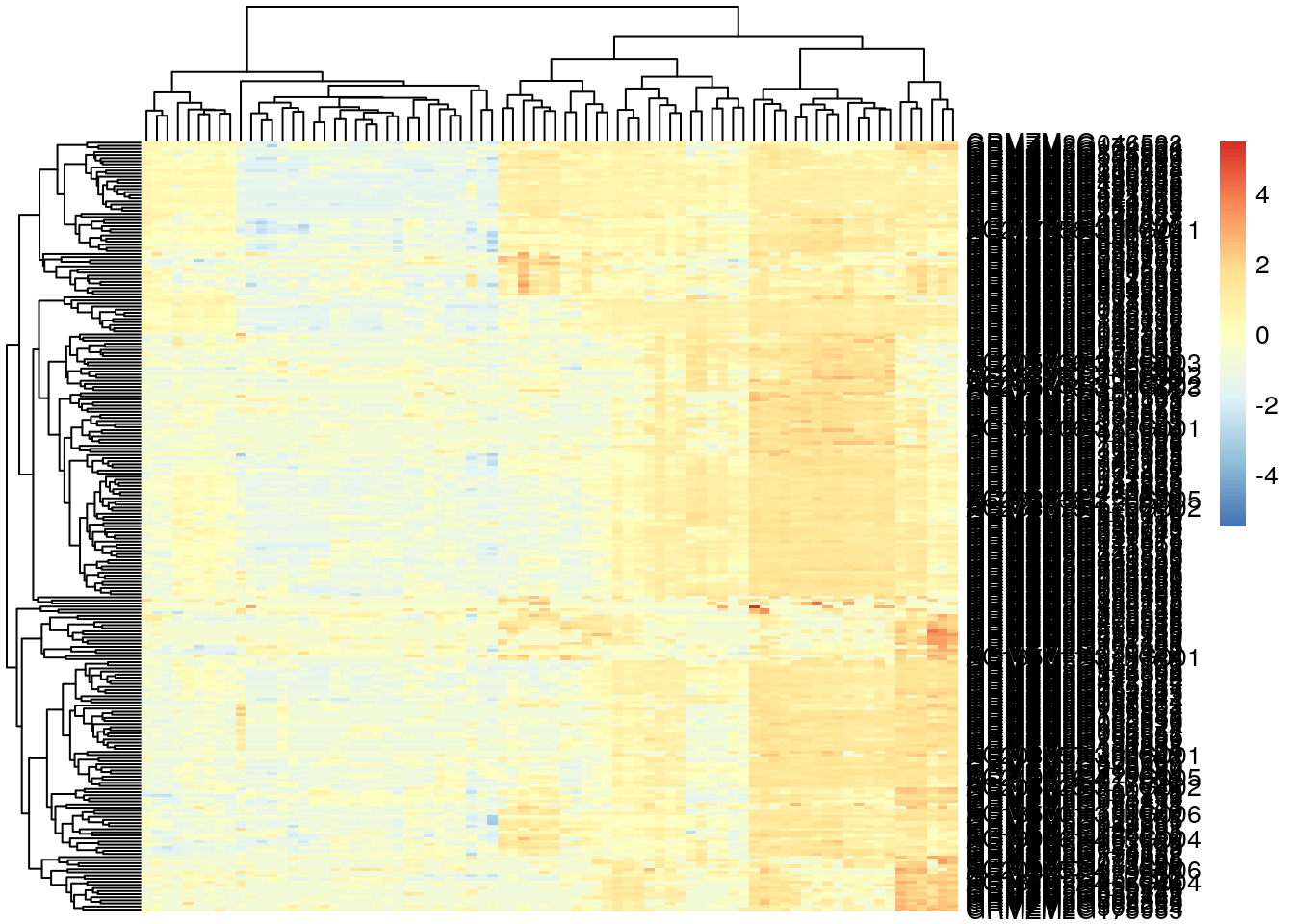
pheatmap(log2(gene_exp + 0.01), scale="row", show_rownames = F, show_colnames = T)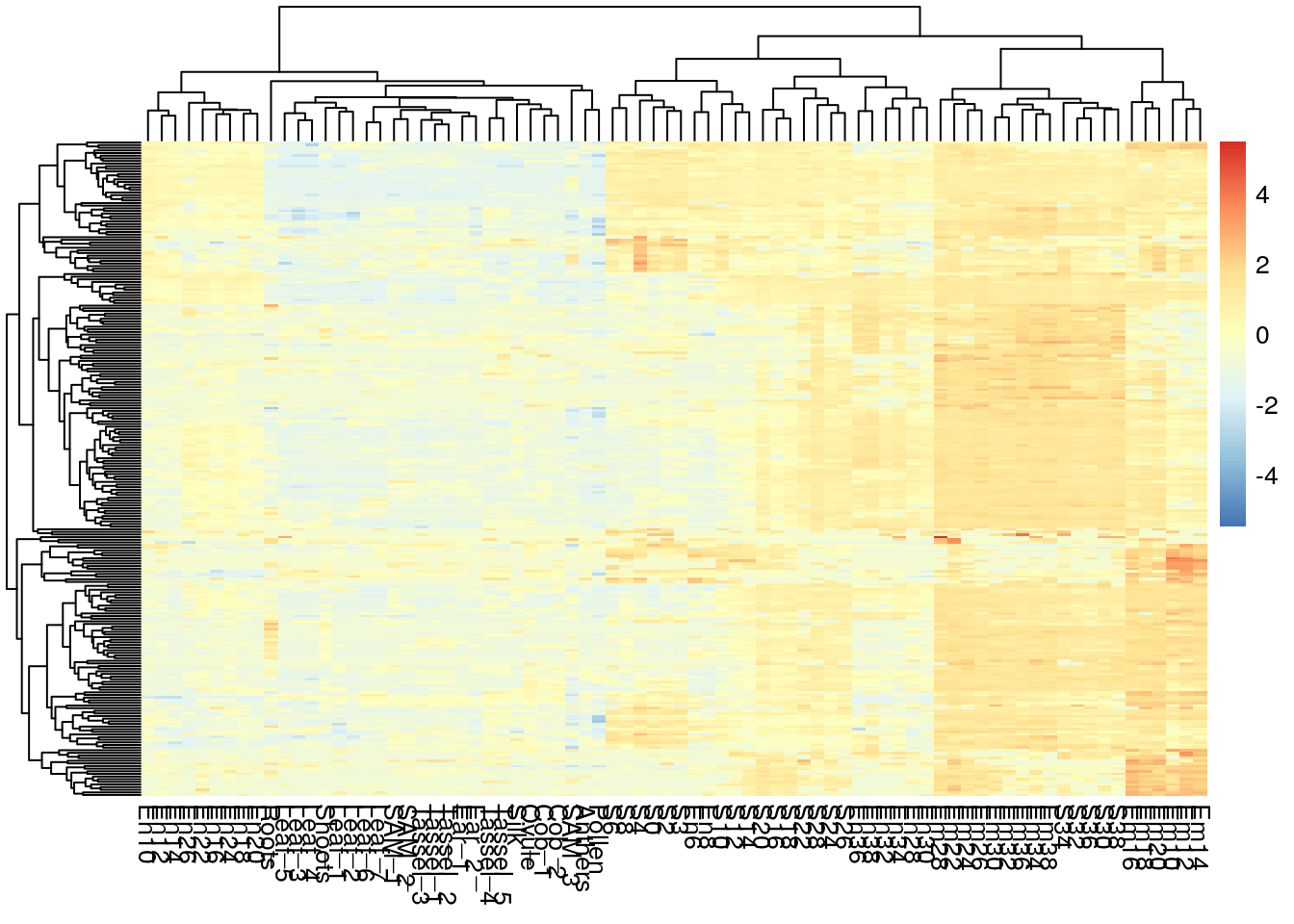
15.3 Add the annotation
15.4
15.5 Transfrom the data
pheatmap(log2(gene_exp + 0.01), show_rownames = F) 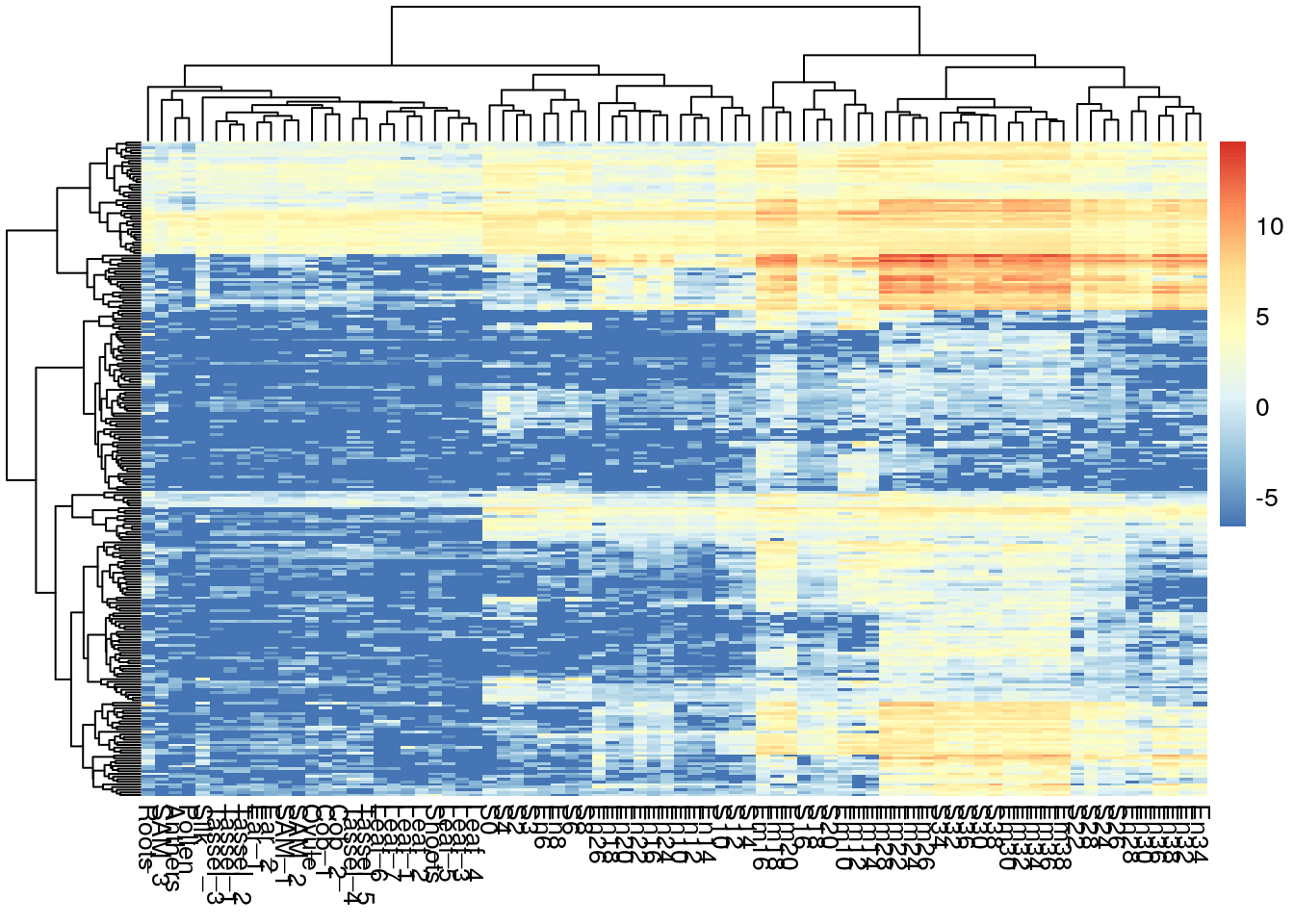
15.6 How to add annotations
15.7 How to cut the trees
15.8 How to get the cluster information from the heatmap
15.9 Change color
15.9.1 Use Brewer
col.pal <- RColorBrewer::brewer.pal(9, "Reds")
col.pal ## [1] "#FFF5F0" "#FEE0D2" "#FCBBA1" "#FC9272" "#FB6A4A" "#EF3B2C" "#CB181D"
## [8] "#A50F15" "#67000D"pheatmap(log2(gene_exp + 0.01), scale="row",
show_rownames = F, show_colnames = T)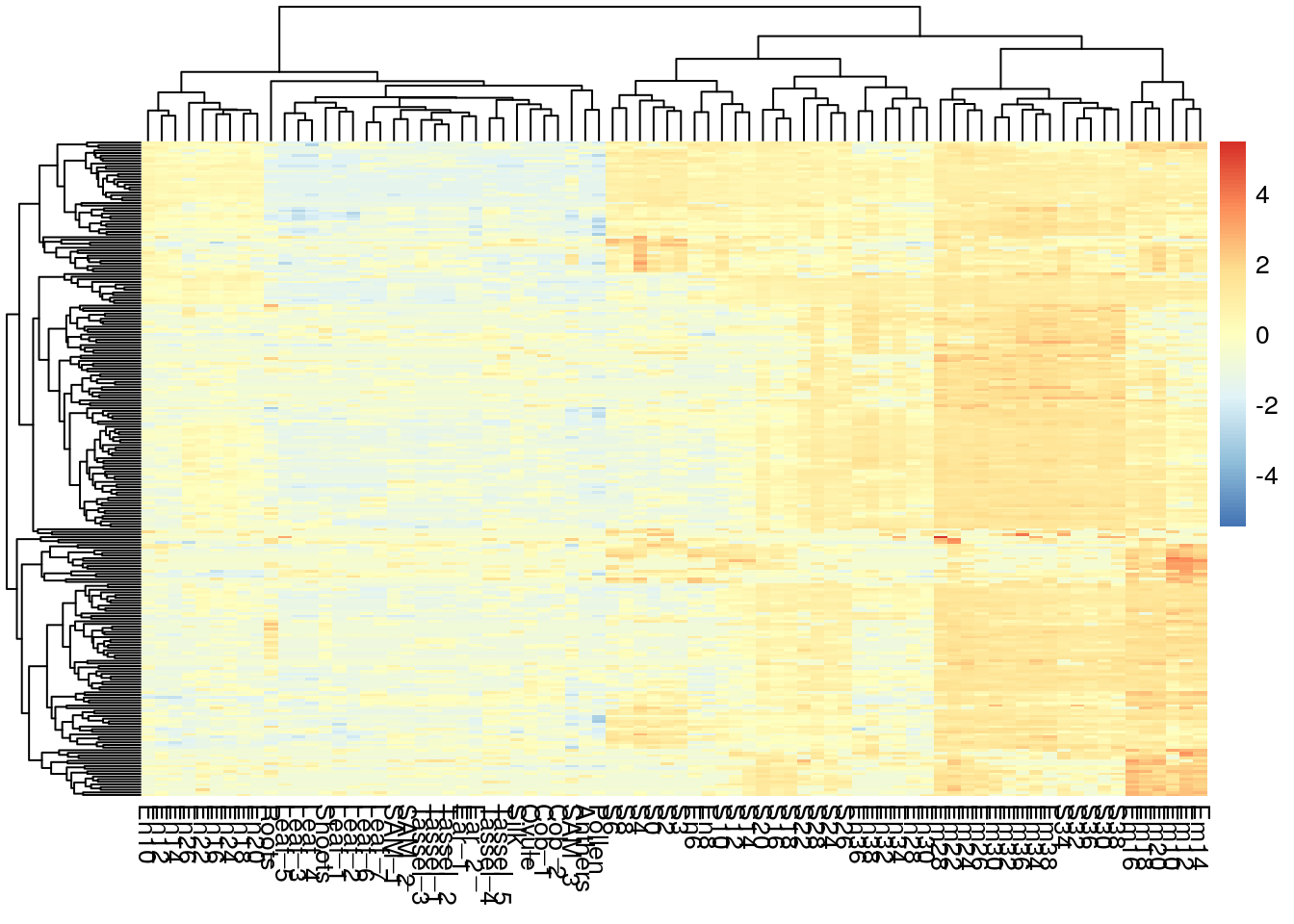
pheatmap(log2(gene_exp + 0.01), scale="row", color = col.pal,
show_rownames = F, show_colnames = T)