Chapter 13 R introduction
I started to work on
13.1 Basic R function
Data structures are variables with informaton stored in. R operates on these data structures. Numberic vector is a single entity consisting of a collection of numbers.
<- is call assignment operator.
# semi-colon (‘;’) can be removed:
gene1_count <- 100;
gene1_count## [1] 100class(gene1_count)## [1] "numeric"gene1_count <- c(100)
gene1_count## [1] 100class(gene1_count)## [1] "numeric"A semi-colon (;) or a newline are used to separate commands
gene_counts <- c(5, 6, 100, 100, 200)
gene_counts## [1] 5 6 100 100 200class(gene_counts)## [1] "numeric"gene1_info <- c(6, "TF")
gene1_info## [1] "6" "TF"class(gene1_info)## [1] "character"13.2 Producing Simple Graphs with R
The credit of this section goes to Dr. Frank McCown (Frank McCown (2006)).
13.2.1 Line Charts
# Define the gene_expr_level vector with 5 values
gene_expr_level <- c(8, 20, 20, 100, 120)
# Graph the gene_expr_fpkm vector with all defaults
plot(gene_expr_level)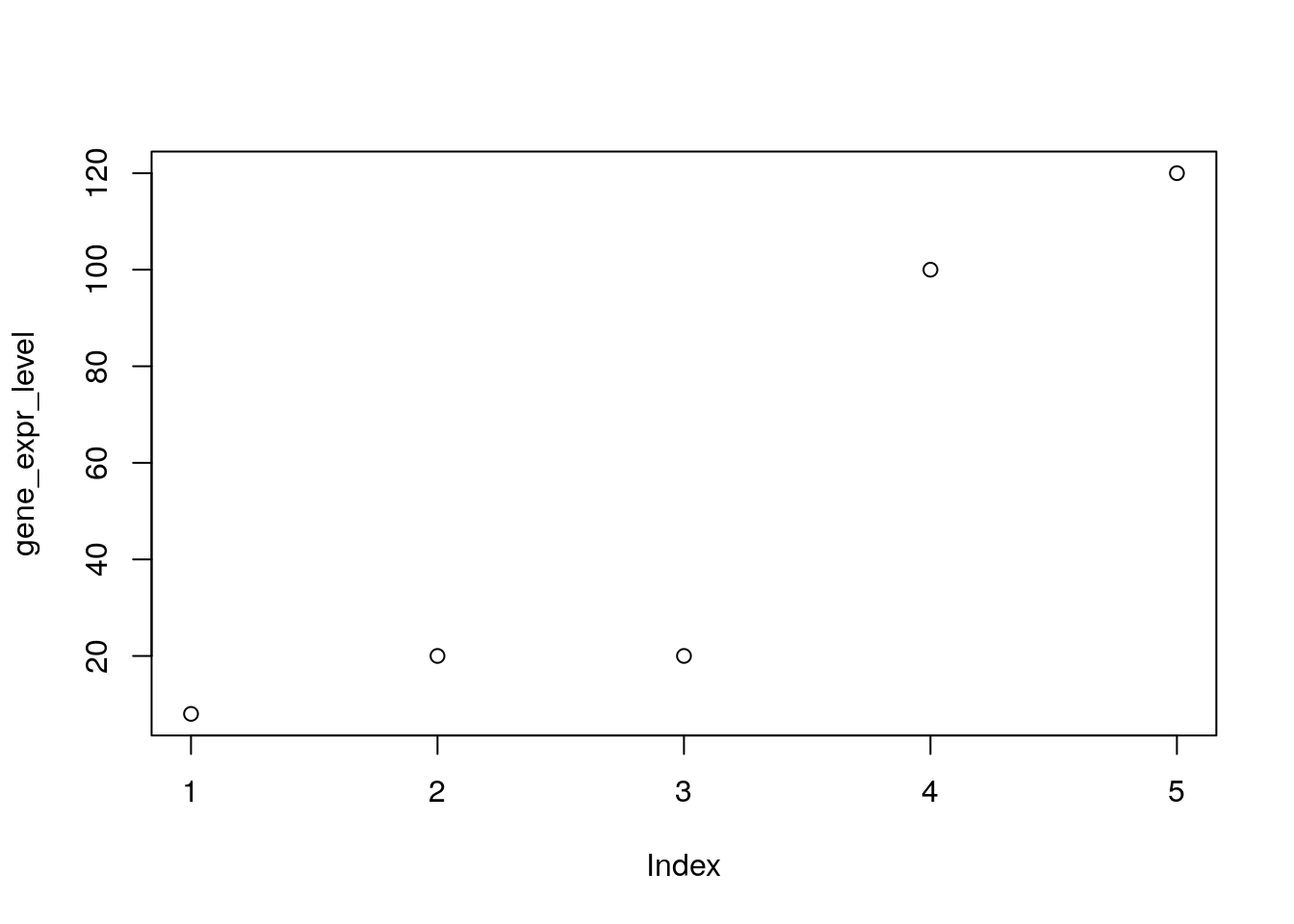
Let’s add a title, a line to connect the points, and some color:
# Define the gene_expr_level vector with 5 values
geneX_expr <- c(8, 20, 20, 100, 120)
# Graph cars using blue points overlayed by a line
plot(geneX_expr, type="o", col="blue")
# Create a title with a red, bold/italic font
title(main="GeneX", col.main="red", font.main=4)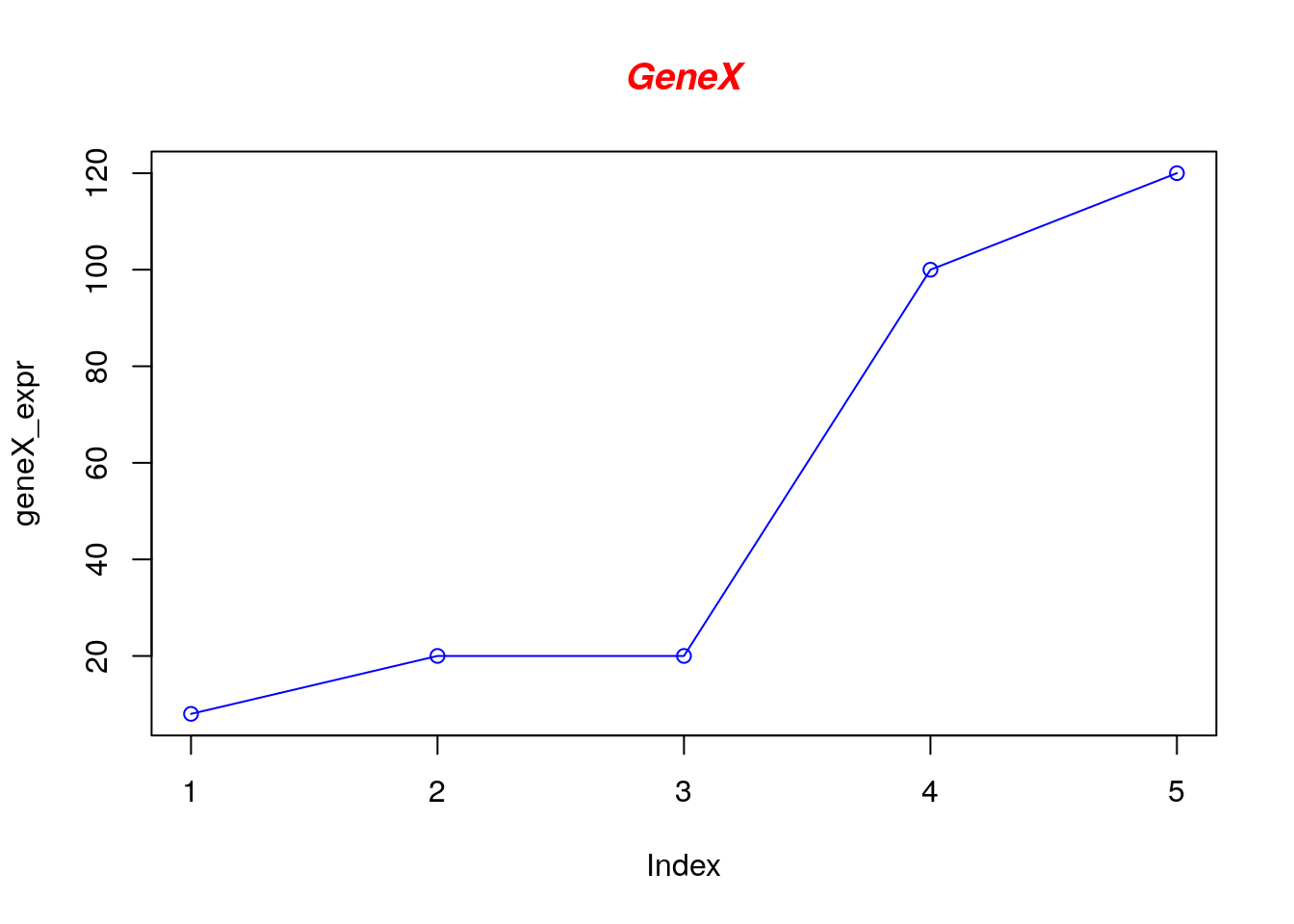
Now let’s add a red line for trucks and specify the y-axis range directly so it will be large enough to fit the truck data:
# Define the gene_expr_level vector with 5 values
geneX_expr <- c(8, 20, 20, 100, 120)
geneY_expr <- c(300, 280, 20, 10, 12)
# Graph cars using blue points overlayed by a line
plot(geneX_expr, type="o", col="blue", ylim=c(0,300))
# Graph trucks with red dashed line and square points
lines(geneY_expr, type="o", pch=22, lty=2, col="red")
# Create a title with a red, bold/italic font
title(main="Gene expresion level", col.main="red", font.main=4)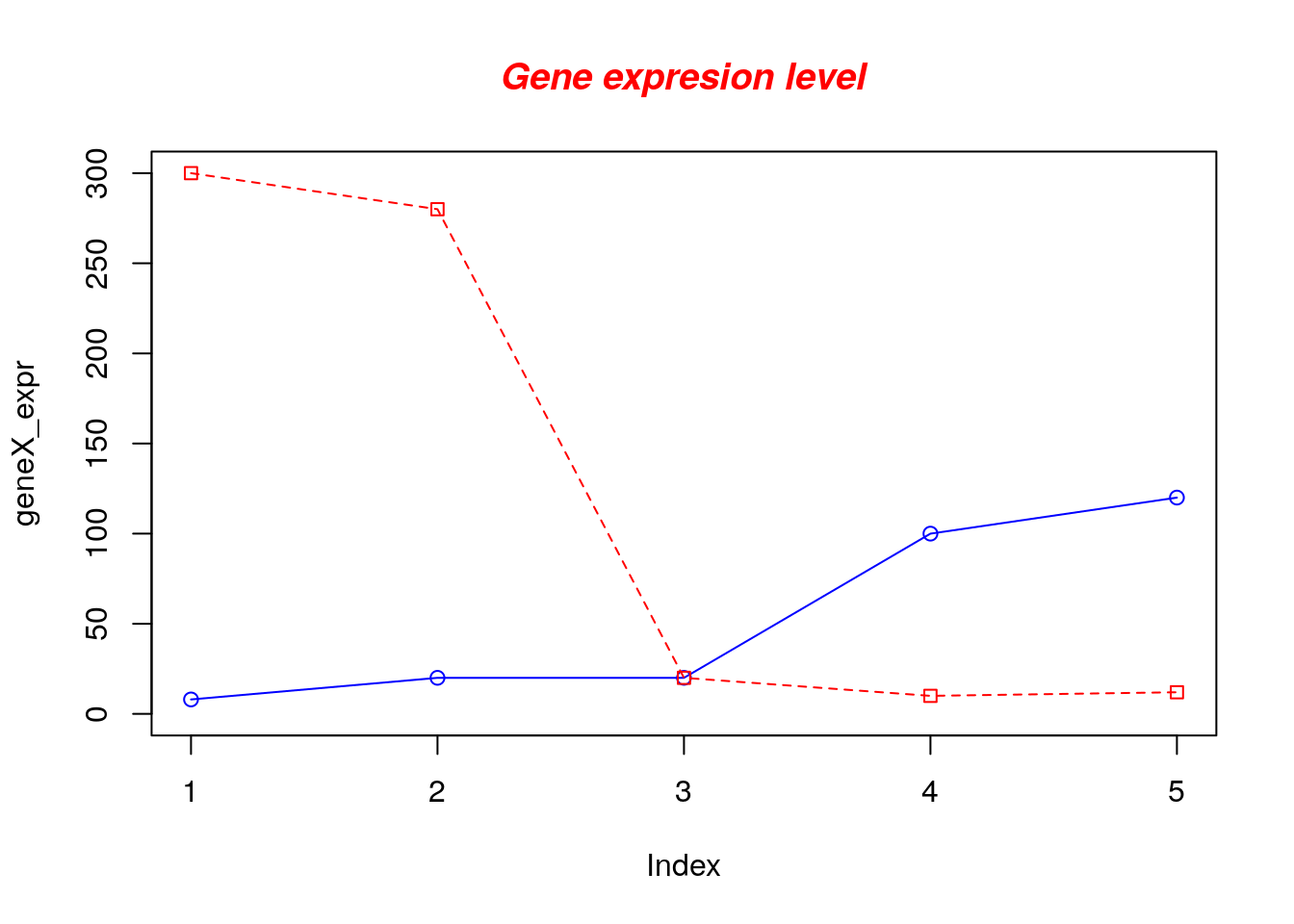
13.3 XXX
fruit = c("apple", "apple", "pear", "orange")
fruit == "apple" ## [1] TRUE TRUE FALSE FALSEfruit = c("apple", "apple", "pear", "orange")
which(fruit == "apple")## [1] 1 2fruit = c("apple", "apple", "pear", "orange")
which(fruit == "apple" | fruit == "pear")## [1] 1 2 313.4 Logic && and |
The short answer is that && and || only ever return a single (scalar, length-1 vector) TRUE or FALSE value, whereas | and & return a vector after doing element-by-element comparisons.
The only place in R you routinely use a scalar TRUE/FALSE value is in the conditional of an if statement, so you’ll often see && or || used in idioms like:
if (length(x) > 0 && any(is.na(x))) { do.something() }
In most other instances you’ll be working with vectors and use & and | instead.
13.5 List as dictionary
the list type is a good approximation. You can use names() on your list to set and retrieve the ‘keys’:
foo <- vector(mode="list", length=3)
names(foo) <- c("tic", "tac", "toe")
foo[[1]] <- 12; foo[[2]] <- 22; foo[[3]] <- 33
foo## $tic
## [1] 12
##
## $tac
## [1] 22
##
## $toe
## [1] 33names(foo)## [1] "tic" "tac" "toe"13.6 Parsing arguments as string
13.6.1 String as xlim
13.6.2 How to access data frame column using variable
a = "col1"
b = "col2"
d = data.frame(a=c(1,2,3),b=c(4,5,6))
colnames(d) <- c("col1", "col2")
d[[a]]## [1] 1 2 3This is useful when you parse a variable from the command line
13.6.3 How to create a formula from a
It can be useful to create a formula from a string. This often occurs in functions where the formula arguments are passed in as strings.
It can be useful to create a formula from a string. This often occurs in functions where the formula arguments are passed in as strings.
design1 = "diet"
design2 = "age"
## `~ diet + age`
as.formula(paste0("~ " , design1, " + ", design2))## ~diet + agecat(readLines('code_R/parse_aug_as.formula.R'), sep = '\n')argv <- commandArgs(trailingOnly = T)
level1 <- argv[1]
level2 <- argv[2]
#First, build a simple data frame with time as a factor and Time as a continuous,
#numeric variable. The two variables look alike when you print the data frame.
#But, if you summarize the data, you see that they are different.
d <- data.frame(level1 = factor(1:4), level2 = 1:4)
colnames(d)<-c(level1, level2)
summary(d)Rscript code_R/parse_aug_as.formula.R time Time## time Time
## 1:1 Min. :1.00
## 2:1 1st Qu.:1.75
## 3:1 Median :2.50
## 4:1 Mean :2.50
## 3rd Qu.:3.25
## Max. :4.00